3.3 Lab
Task: install packages from source#
The default mechanism for installing a package on Linux is to compile from source.
Use the R console in RStudio Workbench to determine your current CRAN repository:
getOption("repos")
Info
You should get the result that the default repository is the RStudio CRAN mirror:
CRAN
"https://cran.rstudio.com/"
attr(,"RStudio")
[1] TRUE
Next, install a package:
install.packages("quantreg", dependencies = TRUE)
Note
Make a note how long this code takes to run.
In addition, note that while the package installs, you get several messages in the console of the form:
* installing *source* package ‘quantreg’ ...
** package ‘quantreg’ successfully unpacked and MD5 sums checked
** libs
f95 -fpic -g -O2 -c boot.f -o boot.o
f95 -fpic -g -O2 -c bound.f -o bound.o
f95 -fpic -g -O2 -c boundc.f -o boundc.o
.
.
<snip>
.
.
f95 -fpic -g -O2 -c srtpai.f -o srtpai.o
gcc -shared -L/opt/R/3.5.1/lib/R/lib -L/usr/local/lib -o quantreg.so boot.o bound.o boundc.o brute.o chlfct.o cholesky.o combos.o crqf.o crqfnb.o dsel05.o etime.o extract.o idmin.o iswap.o kuantiles.o linpack.o mcmb.o penalty.o powell.o quantreg_init.o rls.o rq0.o rq1.o rqbr.o rqfn.o rqfnb.o rqfnc.o rqs.o sakj.o sparskit2.o srqfn.o srqfnc.o srtpai.o -llapack -lblas -lgfortran -lm -lquadmath -lgfortran -lm -lquadmath -L/opt/R/3.5.1/lib/R/lib -lR
installing to /home/jen/R/x86_64-pc-linux-gnu-library/3.5/quantreg/libs
** R
** data
** demo
** inst
** byte-compile and prepare package for lazy loading
** help
*** installing help indices
** building package indices
** installing vignettes
** testing if installed package can be loaded
* DONE (quantreg)
What does all of this mean?
- Those lines starting with
f95 -fpic ...indicates that the Fortran 95 compiler is at work. - And the line starting with
gcc -shared ...indicates that the GNU C compiler (gcc) is at work. - Then you should get some lines starting with
** Rthat tells you the compilation step is complete, and R is now finalizing the package installation. - Once you get
* DONE ...you know that package installation is complete.
Configure RStudio Workbench for binary packages#
You will encounter RStudio Package Manager in a later module in this course. You haven't yet installed RStudio Package Manager, so for now you can use the RStudio demo instance to configure RStudio Workbench.
Identify the binary package installation URL#
You can browse the demo instance at https://packagemanager.rstudio.com
Browse to the setup tab, and find the set of buttons that allow you to choose "Source" or "Binary".
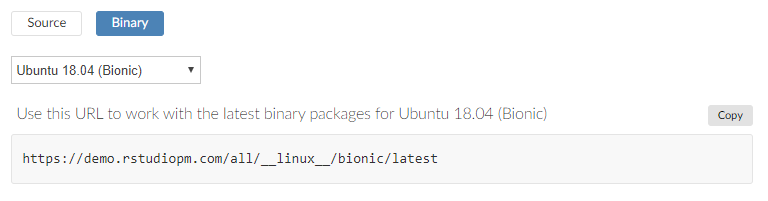
Use the drop-down button to select the correct operating system, then copy the URL
https://packagemanager.rstudio.com/all/__linux__/bionic/latest
Once you have the correct URL, you can configure RStudio Workbench
Task: Configure RStudio Workbench#
You can find the instructions for configuring RStudio Workbench in the RStudio Package Manager admin guide, specifically in section 19.1 A Single Repository.
Read the instructions, then SSH into your VM instance and edit the file
/etc/rstudio/rsession.conf
and add the line
r-cran-repos=https://packagemanager.rstudio.com/all/__linux__/bionic/latest
Save the file, then restart RStudio Workbench:
sudo rstudio-server restart
Also restart your R session
- In RStudio Workbench, you can restart your R session from the menu at
/Session / Restart R, OR - Use the keyboard shortcut
Shift + Ctrl + F10
Task: Test that it works#
Once you've restarted RStudio Workbench, use the R console to check that your repository is correct, and that packages install from binaries without compilation.
To do this, repeat the R commands you used earlier:
getOption("repos")
This time, you should get the URL to the RStudio Package Manager demo site:
CRAN
"https://packagemanager.rstudio.com/all/__linux__/bionic/latest"
Then force the re-install of the quantreg package:
install.packages("quantreg")
This should be the complete output:
Installing package into ‘/home/jen/R/x86_64-pc-linux-gnu-library/3.5’
(as ‘lib’ is unspecified)
trying URL 'https://packagemanager.rstudio.com/all/__linux__/bionic/latest/src/contrib/quantreg_5.54.tar.gz'
Content type 'application/x-gzip' length 1537774 bytes (1.5 MB)
==================================================
downloaded 1.5 MB
* installing *binary* package ‘quantreg’ ...
* DONE (quantreg)
The downloaded source packages are in
‘/tmpr/RtmpsGfCP8/downloaded_packages’
Note that this step is very quick, and there are also no messages from either the fortran compiler (f95) or the C compiler (gcc).
Task: Install the tidyverse#
To speed up the progress of later modules in this course, start the installation of a set of packages you will need later:
install.packages(
pkgs = c(
"tidyverse",
"babynames",
"shiny",
"shinydashboard",
"dbplyr",
"highcharter",
"DT",
"htmltools",
"quantmod",
"dygraphs",
"forecast",
"highcharter",
"odbc",
"plumber",
"ggplot2",
"rmarkdown",
"odbc"
))
This process will take several minutes. Once the installation is running, you can carry on with the rest of the course.